My first-author paper ”Synchrotron X-ray micro-CT as a validation dataset for diffusion MRI in whole mouse brain” has been published in Magnetic Resonance in Medicine. This project has been a huge effort from myself and collaborators at the University of Chicago and Argonne National Laboratory and I’m really excited to see it go out into the world!
Click here to explore our entire microCT dataset online using Neuroglancer.
I published the analysis and visualization code I developed for this project as a Python package called Fiberorient. You can check out the code here.
Highlights
- I developed a 4D spatio-angular registration pipeline between a ~1 TB microCT dataset and a ~100 MB diffusion MRI dataset of the same mouse brain
- I developed computer vision tools to extract local nerve fiber orientations from high-resolution microCT data and bin them as an angular distribution on a basis of spherical harmonic functions
- I demonstrated the feasibility of using microCT as a validation dataset for diffusion MRI
Motivation
Diffusion MRI is sensitive to the diffusion coefficient of water along a given direction. By acquiring 3D images along multiple different directions, the data can be reconstructed to give an estimate of the distribution of nerve fiber orientations at each point in space - a so-called “fiber orientation distribution function” (fODF) . Metrics derived from fODFs have clinical utility in diagnosing diseases that affect white matter integrity. fODFs can also be used to estimate structural connectivity across the brain with fiber tractography.
There are a number of approaches for performing this reconstruction, but all of them require validation with some sort of additional high-resolution imaging because the MRI resolution is not high enough to verify the fiber orientations.
Most studies in the past have used optical microscopy as this validation imaging tool, which works well but is very labor-intensive and requires physically slicing a tissue-sample due to the low penetration depth of optical light. This leads to non-uniform resolution in 3D because the thickness of the slice is typically much thicker than the in-plane resolution of the microscope.
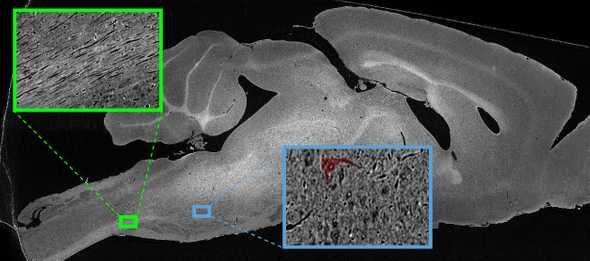
To address this, this paper introduces data from a whole mouse brain collected at the Advanced Photon Source (APS) synchrotron at Argonne National Laboratory. With the powerful x-rays at APS, we can image a whole mouse brain with ~1 micron resolution without having to slice or destroy the sample. This resolution allows us to visualize individual cell bodies and axons:
In the paper, we demonstrate how this type of data can be used to validate diffusion MRI.
Strategy
The primary question we want to answer is: How well does diffusion MRI recover the true orientations of nerve fibers across the brain?
In order to do that, we need to do the following:
- Extract the fiber orientations from the high-resolution microCT data
- Spatially register the two datasets for fair evaluation of diffusion MRI
Feature extraction
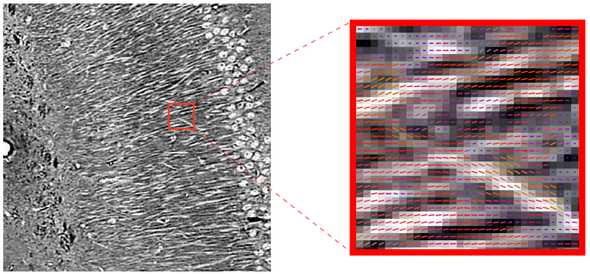
Local fiber orientations were extracted from the microCT data using a computer vision technique called structure tensor analysis. To quickly summarize, structure tensor analysis uses eigenanalysis of local tensors formed by image gradient vectors to estimate the local 3D orientation with the smallest change in intensity value within a predefined neighborhood. We assume that for voxels containing nerve fibers, this orientation points along the main axis of the fiber.
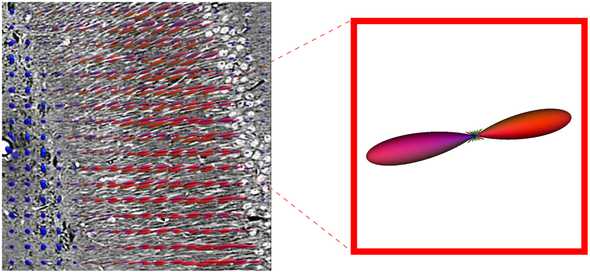
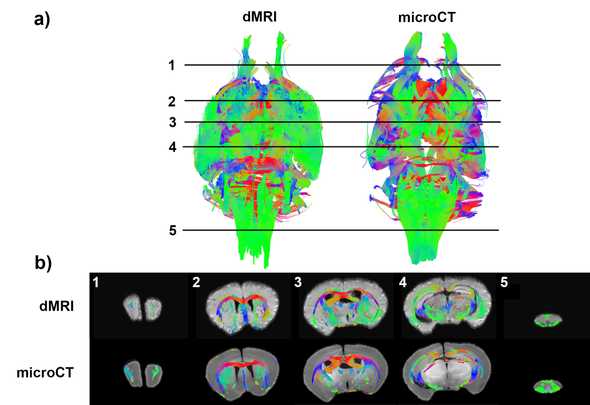
In other words the algorithm takes an image of nerve fibers, and measures the vector of nerve fiber orientations at every pixel, as shown below, where the color of the vector represents direction (left-right: red, up-down: green, in-out: blue).
Now that we know the fiber orientations, we need to find a way to fairly compare them to the low-resolution diffusion MRI results, since there are roughly 2 million microCT voxels for every single MRI voxel. Specifically, we want to bin individual vectors together into an fODF defined on the same scale as the MRI resolution.
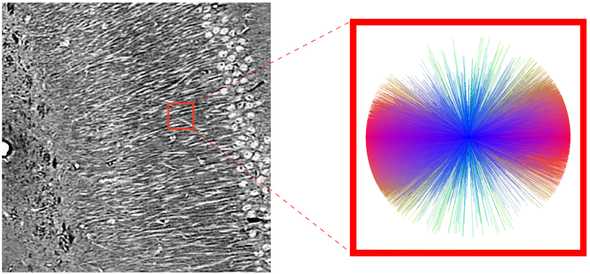
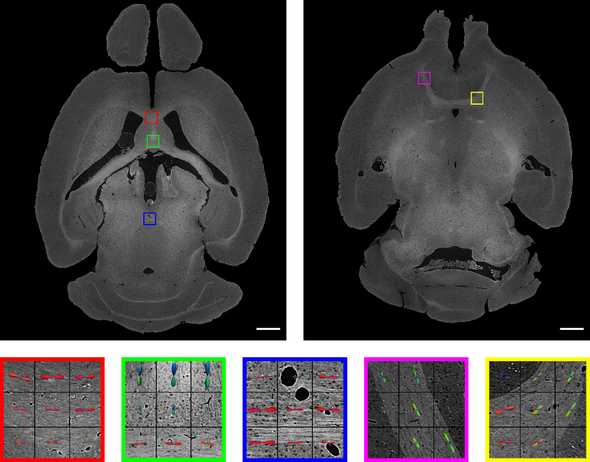
First, we define a 3D cube of space in the microCT data and group all of the vectors within that cube into an angular histogram:
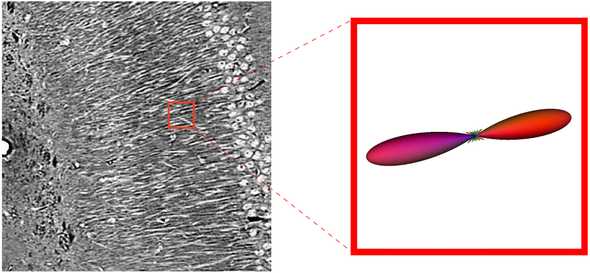
Then, we express our histogram on a basis of spherical harmonic functions to get an fODF. This is the same format of orientation estimate produced by the diffusion MRI reconstruction.
The key is that when we binned the vectors together to make our angular histogram, we chose a 3D cube equal in size to an MRI voxel, so now we have a 3D grid of fODFs on the same scale as the MRI data and can fairly compare them after spatial registration.
See more fODF results from the microCT data below:
Spatial registration
Now that we have fODFs from both modality on the same scale, we need to align the voxel grids so that we can fairly compare individual orientation estimates.
With optical imaging, this registration task is challenging since you are typically dealing with digital stacks of 2D images. The massive advantage of using the whole-brain microCT data with the structure-tensor processing step is that the data is already in the same format as the MRI reconstruction, and the scale difference has already been taken care of, so we can use existing registration packages.
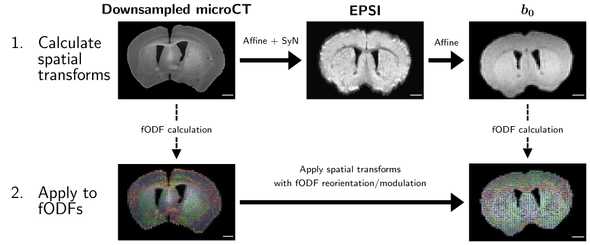
An overview of our registration pipeline is shown below:
To summarize, we first perform purely spatial registrations using the ANTS package:
- We downsample the microCT intensity data and perform nonlinear registration to a structural MRI voxel (EPSI)
- We perform a linear registration from our higher-resolution EPSI data to our low-resolution diffusion data (b0)
We then concatenate both transforms and apply them to the microCT fODFs, using MRTrix3 to account for reorientation and modulation of the fODFs.
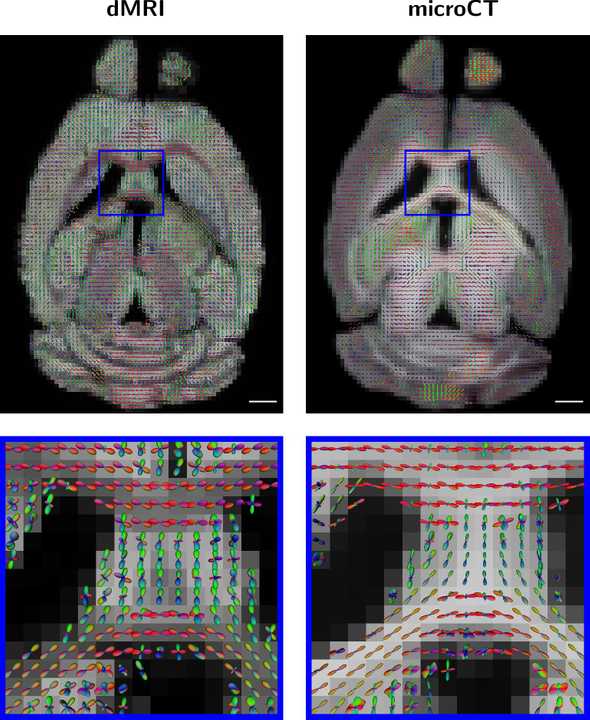
When all is said in done, we get a result like this:
The microCT fODFs were derived from actual computational measurements of the underlying nerve fiber orientations, so they can be used in future studies to benchmark different diffusion MRI approaches.
In another paper, we show how this imaging and registration pipeline can be extended to include nano-scale imaging over small sections with serial electron microscopy.
Tractography
It is extremely difficult to perform whole-brain validation with optical methods due to the issues with 3D alignment I brought up earlier. So another bonus to using microCT is that it allows you to use fiber tractography algorithms on structure-tensor derived fODFs and evaluate how biased fODFs from diffusion MRI affect tractography performance: